Fast5 File Mapping
provided by WunschAG
Drop Fast5 File

or
Drop Fast5 File

or
This webpage accompanies our practical course on CRISPR/Cas gene targeting and nanopore DNA sequencing. Detailed information can be found in our publication. The course was originally developed by Hauke Holtorf, Bernd Dittrich, Thomas Dürr and Jürgen Braun. Röbbe Wünschiers and Robert Maximilian Leidenfrost expanded the course by nanopore DNA sequencing. The following figure illustrates the complete experiment.
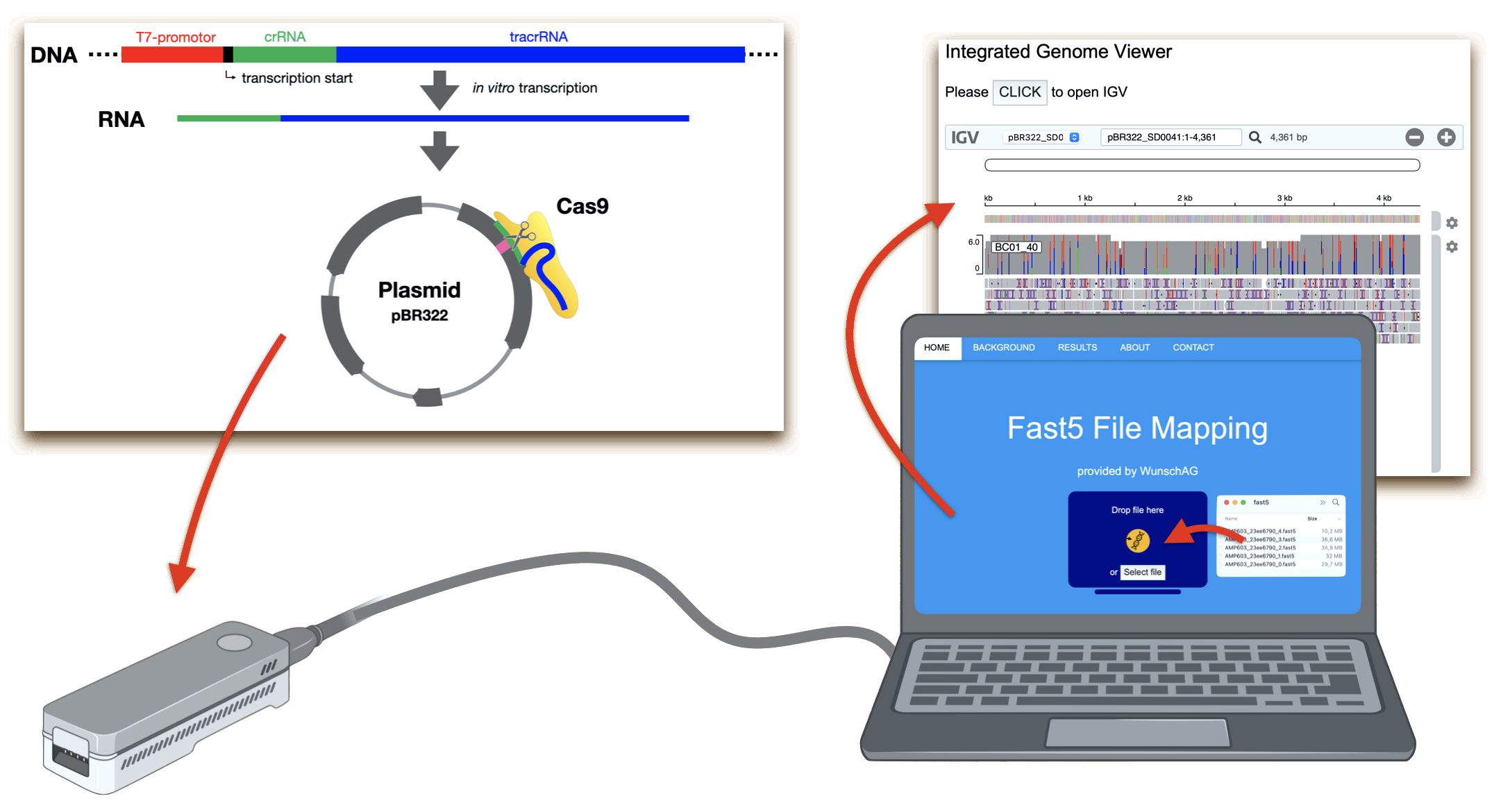
In the laboratory, plasmid DNA is cut by the site specific Cas9 endonuclease. It finds its target with a guide RNA that is previously synthesised from a DNA template. Afterwards, the cut plasmid is sequenced in order to determine the precise cutting site. Although known by the teacher, it is to be determined by the students.
For DNA sequencing the MinION sequencer from Oxford Nanopore is employed. The following video show preparing the MinION equipped with a Flongle flow cell for sequencing.
Finally, the sequence reads produced by the MinION are mapped to the original plasmid and the result is visualised. These bioinformatic steps are executed by our server, accessible through this website. You just have to upload the Fast5 file produced by the sequencer. To test this data analysis pipeline, you can download a demo Fast5 file (please note that file upload is limited to 150 MB).
Take a look at this video to see our experiment in action (german only).
What does this server do? In the background, the guppy software reads a Fast5 file and converts the amperometric measurements (squiggles) into nucleotide reads with the help of a neural network. Note that during sequencing and with standard settings, for every one thousand reads one Fast5 file is created. The first will be available after roughly ten minutes. This base calling process requires quite some time and CPU load. The result is a human-readable FastA file with nucleotide reads. These are split according to their attached barcode sequence by the software porechop and quality filtered with qfilter. Thus, every group can analyse their own sequence read set. The reads are then mapped to the reference, the pBR322 plasmid sequence. This process can be compared to the find-function in a word processor like MS Word. IGV then visualises the result by plotting each read in a line with matching nucleotides one below the other.
Processing might take one hour. Please note that dots are added to indicate that the server is still alive. IGV will open in a new tab after pressing the click button.
In 2012, Jennifer Doudna and Emmanuelle Charpentier co-authored a seminal article in the journal Science describing a programmable, RNA-guided endonuclease1. This enzyme, known as Cas9, leads to targeted double-strand breaks in the DNA2. When repaired by a billion-year-old cellular machinery, the cell can be tricked to insert specific nucleotides or complete genes. While the former allows, e.g., for the repair of genetic diseases, the latter can introduce new functions to cells. Already in 2020, both scientists were awarded the Nobel Prize for developing a genetic engineering tool from what was originally discovered as an adaptive bacterial immune system. One major advantage of this tool, named the CRISPR/Cas gene editing system, is its ease of use: Only a specific guide RNA and the Cas enzyme are required.
In 2018, the Chinese He Jiankui became the first scientist ever who gene-edited human fertilised egg cells3. After successful implantation and birth, two became known as Nana and Lulu – the first twins with a genetically manipulated germ line. Clearly, an ethical boundary was crossed and his experiment outlawed worldwide. Nonetheless, gene editing or, in a broader sense, genome editing does also hold promises to cure people from genetic diseases and to develop, e.g., crops that withstand pests and climatic challenges4.
However, the before said has not been possible without reading the DNA in an efficient and economical way. In the 1970s, Frederick Sanger developed a method to read the DNA sequence with the help of a DNA polymerase5. He received the Nobel Prize in 1980 for what became known as Sanger sequencing. Ever since, DNA sequencing was tedious lab work with devices costing tens of thousands of euros. Until 2014, when the UK-based company Oxford Nanopore Technologies released its first portable sequencing device that employs protein nanopores for DNA sequencing6-7. Their device, named MinION, costs less that one thousand euros and fits into your palm.
Gaining insight into the effect of genetics by reading DNA and associating DNA sequences with phenotypes is the basis for DNA editing, e.g., with the CRISPR/Cas gene editing system. Both methods lay the foundation for future-oriented medicine, breeding, conservation biology or genetic engineering, to name but a few, and ultimately for a modern bioeconomy8.
1Jinek, M. et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816-821 (2012). 2Chan, H. Faster, cheaper, CRISPR: the new gene technology revolution. Science in School 38, (2016). 3Greely, H. T. CRISPR’d babies: human germline genome editing in the ‘He Jiankui affair’. Journal of Law and the Biosciences 6, 111-183 (2019). 4Wünschiers, R. Genes, Genomes and Society (Springer Berlin Heidelberg, Berlin, Heidelberg, 2022). 5Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74, 5463-5467 (1977). 6Deamer, D., Akeson, M. & Branton, D. Three decades of nanopore sequencing. Nat Biotechnol 34, 518-524 (2016). 7Göpfrich, K. & Judge, K. Decoding DNA with a pocket-sized sequencer. Science in School 43, (2018). 8Wang, Y., Zhao, Y., Bollas, A., Wang, Y. & Au, K. F. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol 39, 1348-1365 (2021).
Please site this work as: Wünschiers R, Leidenfrost RM, Holtorf H, Dittrich B, Dürr T, Braun J. 2024. CRISPR/Cas9 gene targeting plus nanopore DNA sequencing with the plasmid pBR322 in the classroom. J Microbiol Biol Educ e00187-23. DOI: 10.1128/jmbe.00187-23
This website stores your Fast5 file and the results.
This website is hosted at the University for Applied Sciences Mittweida. Please read the legal information here.
I like to thank: ► W3Schools for providing nice style classes ► Oxford Nanopore Technologies for their devices, support, guppy basecaller and Fast5 toolkit ► the HDF Group for their HDF5 toolkit ► Ryan Wick for providing porechop ► Heng Li for providing minimap2 ► again Heng Li and others from the Wellcome Trust Sanger Institute for developing samtools ► the igvteam for providing a javascript version of their Integrated Genome Browser ► Fonticons Inc. for providing awesome fonts ► the JS Foundation for providing jQuery.
We try our best to enable your teaching.
Feel free to contact me.